SPRAS Tutorial
Purpose of this tutorial
This tutorial will introduce participants to SPRAS and demonstrate how it can be used to explore biological pathways from omics data.
Together, we will cover:
How to set up and run SPRAS
Running multiple algorithms with different parameters across one datasets
Using the post-analysis tools to evaluate and compare results
Building datasets for analysis
Other things you can do with SPRAS
Prerequisites for this tutorial
Required software:
Conda : for managing environments
Docker : for containerized runs
Git: for cloning the SPRAS repository
A terminal or code editor (VS Code is recommended, but any terminal will work)
(Optional) Cytoscape for visualizing networks (download locally, the web version will not suffice)
Required knowledge:
Ability to run command line operations and modify YAML files.
Basic biology concepts
Note
This tutorial will require downloading approximately 18.3 GB of Docker images and running many Docker containers.
SPRAS does not automatically clean up these containers or images after execution, so users will need to remove them manually if desired.
To stop all running containers: docker stop $(docker ps -a -q)
To remove all stopped containers: docker container prune
To remove unused Docker images: docker image prune
SPRAS Overview
What is pathway reconstruction?
A pathway is a type of graph that describes how different molecules interact with one another for a biological process.
Curated pathway databases provide useful well studied references of pathways but are often general or incomplete. This means they may miss context-specific details relevant to a particular condition or experiment.
Pathway reconstruction algorithms address this by mapping molecules of interest onto large-scale interaction networks (interactomes) to generate candidate context-specific subnetworks that better reflect the high-throughput experimental data.
These algorithms allow researchers to propose computational-backed hypothetical subnetworks that capture the unique characteristics of a given context without having to experimentally test every individual interaction.
Running a single pathway reconstruction algorithm on a single dataset can be challenging, since each algorithm often requires its own input format, software environment, or even a full reimplementation. These challenges only grow when scaling up to using multiple algorithms and datasets.
What is SPRAS?
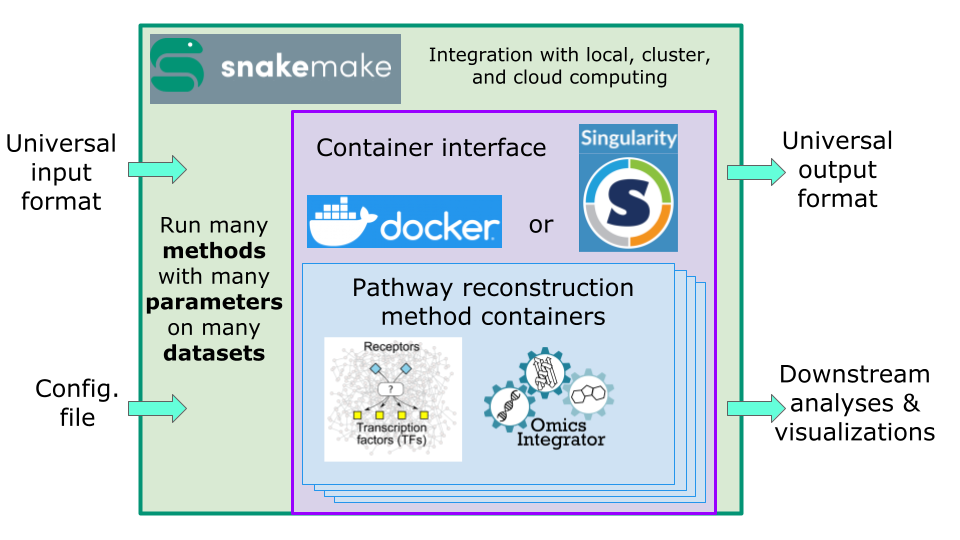
Signaling Pathway Reconstruction Analysis Streamliner (SPRAS) is a computational framework that unifies and simplifies the use of diverse pathway reconstruction algorithms.
SPRAS allows users to run multiple datasets across multiple algorithms and many parameter settings in a single scalable workflow. The framework automatically handles data preprocessing, algorithm execution, and post-processing, allowing users to run multiple algorithms seamlessly without manual setup. Built-in analysis tools enable users to explore, compare, and evaluate reconstructed pathways with ease.
SPRAS is implemented in Python and leverages two technologies for workflow automation:
Snakemake: a workflow management system that defines and executes jobs automatically, removing the need for users to write complex scripts
Docker: runs algorithms and post analysis in a containerized environment.
A key strength of SPRAS is automation. From provided input data and configurations, SPRAS can generate and execute complete workflows without requiring users to write complex scripts. This lowers the barrier to entry, allowing researchers to apply, evaluate, and compare multiple pathway reconstruction algorithms without deep computational expertise.